Detection of Multidrug-Resistant Gram-Negative Bacteria in Untreated Wastewater of Public Hospitals in Northern Iraq
Danyar Hameed M Amin1*, Tara Sedeeq Asaad2, Shanya Baqi Sadiq2, Nizar Hazim Wali2, Tchamou Potindji2
1Department of Biomedical Science, Science, Komar University of Science and Technology, Sulaymaniyah, Iraq
2Department of Medical Microbiology and Clinical Microbiology, Health Science, Near East University, Nicosia, Cyprus
- *Corresponding Author:
- Danyar Hameed M Amin, Department of Biomedical Science, Science, Komar University of Science and Technology, Sulaymaniyah, Iraq; E-mail: Danyar.hameed2@gmail.com
Received date: February 08, 2022, Manuscript No. IPJPHM-22-12973; Editor assigned date: February 11, 2022, PreQC No. IPJPHM-22-12973 (PQ); Reviewed date: February 28, 2022, QC No. IPJPHM-22-12973; Revised date: April 11, 2022, Manuscript No. IPJPHM-22-12973 (R); Published date: April 19, 2022, DOI: 10.36648/IPJPHM.8.4.001
Citation: Amin DHM, Asaad TS, Sadiq SB, Wali NH, Potindji T (2022) Detection of Multidrug-Resistant Gram-Negative Bacteria in Untreated Wastewater of Public Hospitals in Northern Iraq. J Pharm Microbiol Vol:8 No:4
Abstract
Wastewater is considered as a household of antibiotic-resistant Gram-negative pathogens, and access to these microbes to cause a serious health risk. The purpose of this study is to detect Gram-negative bacteria including their antibiotic resistance patterns in the sewer drains of three public hospitals in Ranya-Sulaymaniyah governorate in northern Iraq. The isolates were identified and classified using VITEK-2 automated identification system. Disk diffusion assay was performed to assess antibiotic susceptibility. In total, 732 Gram-negative isolates were identified; Escherichia coli (276/732,37.7%), Pseudomonasaeruginosa (128/732,17.5%), Klebsiella pneumonia (104/732,14.2%), Salmonella entericaserovar Typhi (93/732,12.7%), Shigella sonnei. (72/732,9.8%) and AcinetobacterBaumannii (59/732,8.1%) were identified using conventional method, respectively. Isolates were tested for its susceptibility against a group of antibiotics including; tetracycline, ciprofloxacin, gentamycin, streptomycin, amikacin, ampicillin and azithromycin. Multiple Antibiotic Resistance index (MARI) was used to calculate resistant index, levels of the isolates varied from 0.1 and 0.8%. Escherichia coli was top resistant isolate (237/276, 85.7%) followed by Acinetobacter baumannii (34/59, 57%) to seven antibiotics, other isolates were found to be resistant to at least three tested antibiotics. Resistance was highest to Gentamycin, Amikacin and Tetracycline (57%, for each) and lowest with Azithromycin (14.2%). As a result, multidrug-resistant Gram-negative bacteria were detected in the public hospitals wastewater, which aids their broadcasting infections in the community.
Keywords
Gram-negative bacteria; Antibiotics; Multidrug resistance; Public hospitals; Hospital wastewater
Introduction
Spread of antibiotic resistance as environmental problems and pollutants have largely been detected. Antibiotic resistance is a growing global threat to public health. New resistance mechanisms are gradually arising across the world, posing a challenge to our ability to prevent spread of serious human infections according to European Centre for Disease Prevention and Control (ECDC) [1]. Despite the fact that is a natural occurrence, human and animal in misapply and overuse of antibiotics has intensified the process dramatically [2]. The incidence of resistance to a wide range of antimicrobial agents by a variety of organisms is a major concern facing modern medicine. In fact, not long after the advent of antibiotics as chemotherapeutic agents, resistance emerged [3,4]. According to previous studies, infections caused by antibiotic-resistant species are expected to cause over 10 million deaths globally by 2050, resulting in significant economic damage if the threat is not taken into account [5].
The spread of microorganisms in the community is facilitated by water [6]. Resistance to drugs has, in particular, resulted in detrimental effects such as excessive hospitalization, rising treatment costs, a taxed healthcare system, and much higher death rates. If the issue of antibiotic resistance increases and no successful investigations are adopted, healthcare organizations will face difficulty to choose the best medication to handle infections [7].
Hospital wastewater can be hazardous to public health and ecological balance as it can contain various kinds of pollutants such as radioactive, chemical and pharmaceutical wastes as well as pathogenic microorganisms [8]. Excessive use of antibiotics in agriculture results in spread of resistance genes in environment especially in hospital waste water [9]. Studies have shown that hospital wastewater is a highly selective environment and it contributes to high rate of resistant bacteria discharged into the environment [10]. The occurrence of bacteriophages from samples of animal fecal wastes can be environmental vectors for the horizontal transfer of antibiotic resistance genes [11]. Therefore, characterization of antimicrobial resistance patterns in environmental isolates in an important step towards control and prevention of antibiotic resistance worldwide.
Earlier studies have clearly proofed the correlation between detected Gram-negative bacteria in sewage effluents and hospital infections [12]. Furthermore, antibiotic resistance in microorganisms isolated in areas that allow discharge of antibiotics into wastewater has been reported as well, for instance occurrence of antibiotic resistant superbugs in the receiving river from a pharmaceutical company with a production plant which indicated heavy resistance burden in the sampled isolates [13].
Hospital waste effluents, despite being treated, may contain pathogenic drug-resistant bacteria, which stands as the most dangerous single risk factor for the dissemination of pathogenic and drug resistant organisms into the environment [14]. When a human or an animal is given a drug, from 50% to 90% of it is excreted unchanged. The remainder is excreted in the form of metabolites; chemicals produced as byproducts of the body as well as the interaction with the drug. According to the Centre for Disease Control and Prevention (CDC) estimates, about 22000 tons of antibiotics are produced annually in the United States alone, of which 50% is dispensed to humans. Residues of up to 10 different drugs have been found in such environmental water at concentrations totaling 6 Parts per Billion (ppb). German scientists reported that anywhere from 30 to 60 drugs can be measured in a typical water sample [15]. In a recent study, 58% of samples had at least one antibiotic present while 25% had three or more; sulfamethoxazole, trimethoprim, ciprofloxacin, ofloxacin, lincomycin, and penicillin G [16,17]. Many drugs are also designed to be persistent, so that they can retain their chemical structure long enough to do maintain therapeutic action [18].
Hospital effluent with its high content of multidrug resistant enterobacteria and the presence of enteric pathogens could pose a grave problem for the community [19].
Due to numerous studies on the subject, there seems to be none in Iraq, especially Northern Iraq. As a summary, the objective of this project was to detect the presence of Gram negative bacteria and their resistance patterns from waste water of public hospitals in Ranya, Sulaymaniyah governorate, Iraq.
The aim of the present study is to detect pathogenic Gram-negative isolates including their characterizations of multidrug resistant pattern from wastewater of three public hospitals in Rania, Iraq.
Importance of the study
The public hospital sewages may perform a prominent role in disseminating resistant bacteria especially pathogenic Gram-negative bacteria. Existence of these bacteria in hospitals wastewater displays a threat to public health and unknown magnitude risk to the environment. Presenting the findings is of utmost importance as it directly concerns public health to a regional and even national extent.
Materials and Methods
Sample collection
Samples were collected in three public hospitals. In each hospital 300 mL of wastewater were collected, and directly transported to the laboratory in 2 hours. In the laboratory all of the collected samples were mixed well and prepared for isolation and identification.
Bacterial Isolation and Identification
In order to achieve pure colonies serial dilution of the bacterial suspension was done, 100 μl of the sample dilutions were poured and on nutrient agar plates using spreading method and incubated at 37°C for 24 hours in order to evaluate colony forming unit. MacConkey agar was used for sub-culturing the colonies independently and incubated at 37°C for 24 hours. Bacteriological analysis was used to identify the isolated bacteria including macroscopic examination to examine morphological characteristics using Gram-staining technique based on Bergey's manual of determinative bacteriology guidelines, however, VITEK-2 automated system (bioMerieux) was used to perform standard culture and biochemical characterization to the isolates in order to support with identification [20,21].
Antibiotic susceptibility testing
Antibiotic susceptibility testing was performed on all identified bacterial isolates using disc diffusion assay. The following antibiotic discs were used against Gram negative bacteria; Gentamycin (GN) 10 μg; Ciprofloxacin (CPX) 30 μg; Amikacin (AMK) 30 μg; Ampicillin (AM) 10 μg; Tetracycline (TE) 30 μg and Streptomycin (S) 30 μg. Muller-Hilton agar was applied to culture each detected isolate. After solidification of Muller-Hinton plates at room temperature, suspensions of each tested bacteria were spread onto solid media plates and 0.5 MacFarland of bacteria suspension was spread and antibiotic discs were placed on each media. Then, the inoculated plates were incubated for 24 hrs at 37°C for all tested bacteria. Inhibition zones were expressed in (mm) as the diameter of clear zones around the discs and categorized as resistant (R), Intermediate (I), or sensitive (S) according to (EUCAST) the European Committee on Antimicrobial Susceptibility Testing guidelines [22,23].
Evaluating multiple antibiotic resistance index
Multiple Antibiotic Resistance Index (MARI) percentage was measured through equation:
MARI=a/b x 100
“a” indicates the antibiotics that have been successfully resisted by detected Gram negative bacteria in the current study while “b” represents the overall antibiotics used in the test [24].
Results
Bacterial count of water samples
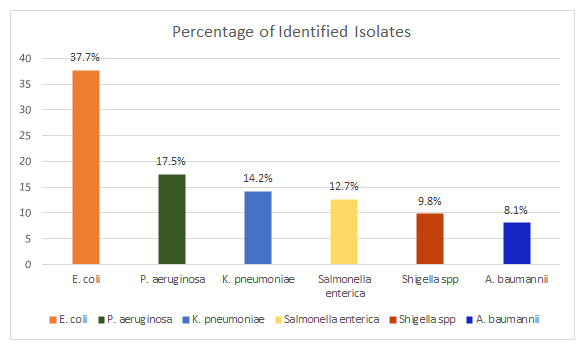
Colony Forming Units (CFU) resulted to range from 1.99*103/ml to 3.27*103/ml colonies. Among 900 mL of collected wastewater from the three hospitals 732 Gram-negative colonies were identified in the current study, bacteriological characterization showed the following; Escherichia coli (276/732,37.7%), Pseudomonas aeruginosa (128/732,17.5%), Klebsiella pneumoniae (104/732,14.2%), Salmonella enterica serovar Typhi (93/732,12.7%), Shigella sonnei (72/732,9.8%), Acinetobacter Baumannii (59/732, 8.1%) (Figure 1).
Antibiotic susceptibility test
The resistance profile to the seven antibiotics was performed to all Gram-negative isolates. Among 732 isolates, most of those isolates were resistant to gentamycin, amikacin and tetracycline; however, E. coli (237/276,85.7%) and (34/59,57%) A. baumannii displayed the highest degree of resistance to all antibiotics, followed by S. enterica serovarTyphi (40/93, 42.8%), K. pneumoniae (45/104,42.8%) and P. aeruginosa (55/128,42.8%), the least resistance pattern was belonged to Shigella sonnei (10/72,14.2%). Antibiogram resistance was calculated to evaluate the drug's efficiency against the identified Gram-negatives. Resistance to GN, AMK and TE (57% for each) was highest, while resistance to AZM was the lowest (14.2%).
The result of antibiotic susceptibility test was evaluated by measuring the clear zone around each antibiotic disc to determine zone of inhibition (Table 1). According to EUCAST the isolates were categorized into Resistant (R), Intermediate (I) and Susceptible (S) [25]. The resistance distribution among the isolates and their zone of inhibition where as follows:
Table 1: Zone of inhibition (mm) and resistance pattern.
| GN mm | CPX mm | AMK mm | AMP mm | TE mm | AZM mm | S mm | |
|---|---|---|---|---|---|---|---|
| E. coli | R (11) | R (14) | R (12) | R (10) | R (13) | S (25) | R (13) |
| P. aeruginosa | S (23) | S (24) | S (27) | R (13) | I (16) | R (11) | R (10) |
| K. pneumoniae | R (10) | S (27) | R (12) | I (15) | R (13) | S (24) | S (26) |
| S. enterica serovar Typhi | R (14) | S (22) | R (13) | I (15) | R (12) | S (22) | I (16) |
| Shigella sonnei | S (26) | S (25) | S (21) | I (17) | R (11) | S (23) | S (24) |
| A. baumannii | R (12) | R (13) | R (10) | R (11) | I (14) | S (22) | I (15) |
*GN:Gentamycin; CIP:Ciprofloxacin; AMK:Amikacin; AMP:Ampicillin; TE:Tetracycline; AZM: Azithromycin; S:Streptomycin.
Evaluation of Multiple Antibiotic Resistance Index (MARI)
Multiple antibiotic resistant bacteria are present in isolates with a MARI value of 0.3>, indicating the existence of highly resistant bacteria. The level of MARI identified isolates varied between 0.1 and 0.8 percent. Each Gram-negative isolate with three or more antibiotic resistance phenotype was recorded as multidrug resistance pathogen according to MAR phenotype assay. Majority of the pathogens were resistant to more than three antibiotics, excepting Shigella sonnei. Basically, isolates with resistance category to three or more antibiotics will be recorded as multidrug resistance MDR, in the current study resulted to have five (5) identified Gram-negative bacteria which had resistance to 3 to 6 antibiotics (Table 2).
Table 2: Distribution of antibiotics resistance among bacteria obtained from sewage drains of the three public hospitals.
| Identified Isolates | List of antibiotics | Number of antibiotics | MARI |
|---|---|---|---|
| E. coli | GN, CPX, AMK, AM, TE, S | 6 | 0.8 |
| P. aeruginosa | AMP, AZM, S | 3 | 0.4 |
| K. pneumoniae | GN, AMK, TE | 3 | 0.4 |
| S. enterica serovar Typhi | GN, AMK, TE | 3 | 0.4 |
| Shigella sonnei | TE | 1 | 0.1 |
| A. baumannii | GN, CPX, AMK, AM | 4 | 0.6 |
The most significant percentages of resistance in the Gram-negative Bacterial isolates were observed in gentamycin, amikacin and tetracycline (57% for each) followed by ampicillin (42.8%). Lower percentages of resistance were found for ciprofloxacin and streptomycin (28.6%), and the lowest resistance to azithromycin (14.2%).
Discussion
Multidrug resistance MDR Gram-negative bacteria considered as a significant problem to public health. The resistance pattern of Gram-negative bacteria has previously been investigated. This study focuses on Gram-negative bacteria found in hospital sewages and their resistance to the most widely used antibiotics in the area. Hospital infections have been correlated with Gram-negative bacteria in previous research and have been found in sewages [26]. In the current study, sewage drains from three hospital units were tested for detecting the prevalence of Gram-negative bacteria including their resistance strains. Findings showed a high bacterial percentage in the sewages; the main cause is that bacteria get accumulated in the sewages and achieve optimal environment for growth.
As a consequence, allowing human waste containing acquired resistant pathogens to enter large water sources without being adequately controlled may promote the spread of bacterial resistance isolates. There are also previous cases of high bacterial counts in hospital waste and effluents [27]. Micro and macro pollutants are found in hospital wastewaters from a variety of sources, including operating rooms, laboratories, investigation units, polyclinics, and drug use sectors. Bacteria and viruses are the most significant macro pollutants, while antibiotics, heavy metals, hormones, and detergents/antiseptics are the most significant micro polluters [28].
Conversely, experiments have been identified in related conditions that enable the discharge of antibiotic wastes into receiving water sources, such as the incidence of antibiotic-resistant superbugs from pharma companies, which revealed a high degree of resistance in the isolates [29].
The results of this study, which tested five deferent classes of antibiotics such as fluoroquinolone, aminoglycoside, tetracycline, macrolide, and penicillin antibiotics and seven antibiotics including Tetracycline, Gentamycin, Streptomycin, Ciprofloxacin, Ampicillin, Amikacin and Azithromycin have been used against six types of identified Gram-negative bacteria which considered to determine the amount of antibiotic resistance in northern Iraq, are presented in Table 1.
Bacteriological analyses of sewage water from investigation sites revealed that among Gram-negative bacteria isolated, enteric pathogens are the most predominant subgroup. In fact, this group accounted for almost 75% of all isolates; among them, E. coli was the most prevalent bacteria, accounting for n=276 isolates (37.7%), followed by K. pneumoniae, S. enterica serovar typhi and S. sonnei (respectively 14.2%, 12.7% and 9.8% of the total isolates tested). The high isolation rate of enteric bacteria from sewage water is in agreement with previously reported observations from wastewater analyses, including hospital sewage [30].
In the current absence of any kind of wastewater treatment plant in all investigated healthcare facilities, the disposal of untreated hospital wastewater containing antibiotic-resistant bacteria may result in a greater likelihood of waterborne enteric disease outbreak caused by the identified enteric pathogens. In fact, Salmonella spp has already been reported in Thi-Quar region, as a cause of gastroenteritis in children [31]. Of note, and in accordance with the hypothesis of hospital wastewater as possible source of environmental pollution. made a relevant finding when they detected two identical K. pneumoniae respectively originated from hospital sewage and urban wastewater [32]. Other than enterobacteria, P. aeruginosa and A. baumannii which have recently emerged as leading cause of hospital acquired infections were also recovered from tested samples and respectively accounted for 17.5% and 8.1% of the isolates.
Hospitals and more generally healthcare facilities are settings where antibiotics are extensively used and where resistant bacteria may thrive selectively and therefore consistently being discharged in related wastes [33]. Considering the World Health Organization priority to address antibiotic resistance in pathogenic bacteria and taking in account the antibiotics mostly used in clinical practice, this study assessed the antimicrobial resistance profile to antibiotics from four classes: fluoroquinolones (ciprofloxacin), aminoglycosides (gentamycin, streptomycin and amikacin), tetracycline’s (tetracycline), macrolides (azithromycin) and penicillin’s (ampicillin).
In the present study, E. coli was found to be resistant to all antibiotics except azithromycin. The extended resistance profile observed in this study is in accordance with results presented in several studies; while one report revealed multi-drug resistance pattern in E. coli isolates, a review from Fouz et al. generalized the findings to several bacterial species [34,35]. In 2019, one study also revealed pan resistance to all antibiotic tested toward E. coli [36]. One could possibly infer that such resistance seen in E. coli is directly due to the continuous discharge of those antibiotics in waste water; accordingly, authors have revealed the strong connection between hospital wastewater and prevalence of multi resistant E. coli [37]. Antimicrobial resistance in Acinetobacter baumannii was also predominant; in fact, it is the second most resistant bacterial isolate.
Also discussing drug susceptibility, one study focused on compared-analyses of drug resistance in bacteria from both treated and untreated wastewater clearly demonstrated lower level of resistance in treated water isolates. Similarly, to our results, the latter study presented high level of resistance to ampicillin and tetracycline, whereas, lower resistance was seen in gentamycin (conversely to our findings) [38].
The above presented findings directly concerns public health to a regional and even national extent. Indeed, our investigation, backed up with previous literature have led to the conclusion that absence of wastewater treatment settings in hospitals could contribute to a larger environmental hazard, through dissemination of antibiotic resistance and also influence antibiotic stewardship [39]. Although Asfaw et al. have observed lower frequency of bacterial isolation in treated hospital waste water compared to untreated one, studies conducted in Iraq concluded that sewages, even treated still contain residual pollutants making them unsafe to be discharged in larger water bodies [40,41]. Without any ready-to-use practice for hospital sewage management, one could refer to our study however suffers from some limitations. Although assumed to be the most prescribed and used antibiotics at the time of the study, no attempt had been made toward assessing the concentration of the tested antibiotics in sewage. Thus, we could only assume that observed resistance was due to discharged antibiotics. Further studies are needed to understand how hospital sewage discharge influences larger water-receiving bodies.
Another research reported the existence of antibiotic-resistant bacteria in hospital drainage, raising concerns regarding wastewater treatment's effectiveness in removing antibiotic-resistant bacteria. This poses a significant risk to public health and can contribute to the prevalence of antibiotic resistance [42].
In the previous studies, E. coli, P. aeruginosa and K. pneumoniae presented higher frequency of isolation, and microbial profile of hospital effluents was confirmed the incidence [43,44]. The microbial composition of sewage differs based on local actions, although the majority of sewage microbial composition comes from the gastrointestinal tract, and most bacteria detected in this analysis can be contained in human intestinal microbiota, as well as pathogens, from this analysis [45]. In this respect, E. coli is the most frequent cause of urinary infections and the presence of the same pathogens in inpatients were reported previously.
This study evaluated five deferent classes of antibiotics against three identified Gram-negative families, as shown E. coli under family Enterobacteriaceae is the highest resistance to four groups of antibiotics, more specifically; aminoglycoside (gentamycin), fluoroquinolones (ciprofloxacin), penicillin (ampicillin) and tetracycline, but it is sensitive to macrolide (azithromycin) which almost all of the isolates are sensitive to and it except P. aeruginosa. However, P. aeruginosa resulted to have resistant strains to penicillin (ampicillin), macrolide (azithromycin) and aminoglycoside (streptomycin) and they showed sensitive to tetracycline, gentamycin, amikacin and ciprofloxacin. Regarding S.enterica serovar Typhi and K. pneumoniae, they both had resistance to the same antibiotics namely, gentamicin, amikacin and tetracycline and two of them were sensitive to the other antibiotics. Based on the result, Acinetobacter Baumannii is the second highest resistance after E. coli; it has resistance to three classes of antibiotics such as aminoglycoside (gentamycin and amikacin), fluoroquinolone (ciprofloxacin) and penicillin (ampicillin).
Studies of bacterial resistance profiles linked to hospital sewage sources have been recorded in Nigeria's southern and northern regions [46,47]. According to antimicrobial resistance testing in previous studies, a large number of bacteria have become resistant to ampicillin and amoxicillin. Ceftazidime, a broad-spectrum cephalosporin used to treat Gram-negative infections. Previous studies revealed common bacterial resistance to a variety of antibiotic classes [48]. This may be related to the epidemiological profile of pathogens and the antibiotic treatment protocols used in various countries. The importance of controlling susceptibility patterns in hospital sewage to ensure the proper antibiotic use and prevent resistance mechanisms from expanding into the community is demonstrated mostly by previous studies.
Eventually, since resistance determinants transmit quickly in aquatic matrices, the sewage of public hospitals found to have a high occurrence of MDR bacteria.
Antibiotic resistance is caused by inappropriate usage, misuse, and overuse of antibiotics. As a result, information about antibiotic use should be provided, and unnecessary and excessive use should be avoided. Antibiotic‐resistant bacteria are becoming viral and increasing burden on healthcare and health organizations. Antibiotic‐resistant is a major problem around the world, according to the American Centers for Disease Control and Prevention and the Food and Agriculture Organization. Acquired resistance bacteria predicted to be the death cause of 700,000 people annually. Consequently, bacterial infections and their diseases will be widened due to temperature change, rainfall pattern and climate sensitive bacteria. Antibiotic misuse has a directly and indirectly impact on human health and affect the treatment of fish infections. Fish bacteria and pathogenic fish bacteria acquire resistance as a result of it. Whenever resistant strains of bacteria cause disease, they cause chronic diseases. The transfer of resistance plasmids from bacteria to human diseases is one example of indirect antibiotic resilience. Human microorganisms that significantly contribute resistance through this way cause resistant diseases in humans. Multiple antibiotic resistance genes are also passed from fish borne diseases to human infection, according to research in China [49].
Infections caused by MDR pathogens are correlated to higher rates common or fatal bacterial infections, respiratory tract infections, gastro-intestinal infections, urinary tract infections UTIs, skin and soft tissue infections SSTIs, bacteremia followed by other threatening diseases.
Antibiotics are used in both people and animals to control and treat diseases. This could result in prevailing of antibiotic resistant bacteria in the community. In order to prevent the increasing prevalence of antibiotic-resistant bacteria, people's consciousness of antibiotic use must first be raised. Antibiotics should be used less frequently in both human and animal treatment. All types of certain water among all wastewater effluents, particularly hospital sewages, should be pretreated such as mechanical, chemical, and thermal treatment techniques before being sent to biological wastewater treatment plants.
Conclusion
Prevalence of multidrug-resistant Gram-negative bacteria was found to have a high percentage among public hospitals sewage in the current study. Furthermore, it delivers recent pattern of antibiotic resistance in hospital wastewater and raises concerns regarding the spread of multidrug resistance in the public health system and eventually expanded into wider water sources. The findings showed an identical result to those covered in other regions of the world, and recommends farther examination regarding molecular investigation and control of antibiotic resistance by providing new awareness on the resistance pattern in public hospitals wastewater in Ranya, Sulaymaniyah governorate in northern Iraq.
Availability of data and materials
The dataset used and analyzed during the study are available from the corresponding.
Competing Interests
The authors declare of having no competing interest.
Funding
The research was self-funded project.
Authors Contributions
DHM, TSA, SHBS, NHW and TP participated in the study design, sampling, performing laboratory work and statical analysis. DHMA supervised the study. All authors read and approve the manuscript.
Acknowledgement
We would like to thank Salam Laboratory and Pharmacy Plus Laboratory which are private laboratories for their contributions to perform the practical work of the project.
References
- Magiorakos A, Srinivasan A, Carey R, Falagas ME, Falagas M, et al. (2012) "Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance”. Clin Microbiol Infect 18: 268-281
[Crossref] [Google Scholar] [Indexed]
- Hiller C, Hübner U, Fajnorova S, Schwartz T, Drewes J (2019) "Antibiotic Microbial Resistance (AMR) removal efficiencies by conventional and advanced wastewater treatment processes: A review." Sci Total Environ 685: 596-608
[Crossref] [Google Scholar] [Indexed]
- Jasovský D, Littmann J, Zorzet A, Cars O (2016) "Antimicrobial resistance-a threat to the world's sustainable development." Ups J Med Sci 121: 159-164
[Crossref] [Google Scholar] [Indexed]
- Tacconelli E, Sifakis F, Harbarth S, Schrijver R, van Mourik M, et al. (2018) "Surveillance for control of antimicrobial resistance." The Lancet 18: 99-106
[Crossref] [Google Scholar] [Indexed]
- De Kraker M, Stewardson A, Harbarth S (2016) "Will 10 million people die a year due to antimicrobial resistance by 2050?." PLoS med 13:1002184
[Crossref] [Google Scholar] [Indexed]
- Amaya E, Reyes D, Paniagua M, Calderon S, Rashid M, et al. (2012) "Antibiotic resistance patterns of Escherichia coli isolates from different aquatic environmental sources in León, Nicaragua." Clin Microbiol Infect 18:347-354
[Crossref] [Google Scholar] [Indexed]
- Shrivastava SR, Shrivastava PS, Ramasamy J (2018) "World health organization releases global priority list of antibiotic-resistant bacteria to guide research, discovery, and development of new antibiotics." J Med Soc 32:76-77
[Crossref] [Google Scholar] [Indexed]
- PPCP and Characterising (2013) "High on pollution: drugs as environmental contaminants." J Environ Monit 5:42-46
[Crossref] [Google Scholar] [Indexed]
- Iversen A, Kühn I, Franklin A, Möllby R (2002) "High prevalence of vancomycin-resistant enterococci in Swedish sewage." Appl Environ Microbiol 68:2838-2842
[Crossref] [Google Scholar] [Indexed]
- Yang C, Lin M, Liao P, Yeh H, Chang B, et al. (2009) "Comparison of antimicrobial resistance patterns between clinical and sewage isolates in a regional hospital in Taiwan." Lett Appl Microbiol 48: 560-565
[Crossref] [Google Scholar] [Indexed]
- Schwarz S, Feßler AT, Loncaric I, Wu C, Kadlec K, et al. (2018) "Antimicrobial resistance among Staphylococci of animal origin." Microbiol Spectr 6
[Crossref] [Google Scholar] [Indexed]
- Schwartz T, Kohnen W, Jansen B, Obst U (2003) "Detection of antibiotic-resistant bacteria and their resistance genes in wastewater, surface water, and drinking water biofilms." FEMS Microbiol Ecol 43: 325-335
[Crossref] [Google Scholar] [Indexed]
- Li D, Yu T, Zhang Y, Yang M, Li Z, et al. (2010) "Antibiotic resistance characteristics of environmental bacteria from an oxytetracycline production wastewater treatment plant and the receiving river." Appl Environ Microbiol 76: 3444-3451
[Crossref] [Google Scholar] [Indexed]
- Daniel DS, Lee SM, Dykes GA, Rahman S (2015) "Public health risks of multiple-drug-resistant Enterococcus spp. in Southeast Asia." Appl Environ Microbiol 81: 6090-6097
[Crossref] [Google Scholar] [Indexed]
- Haque, Mainul (2019) "Antibiotic Use, antibiotic resistance, and antibiotic stewardship--A globalpublic consequences." J med sci 18:169-170
[Crossref] [Google Scholar] [Indexed]
- Brown SD, Rybak MJ (2004) "Antimicrobial susceptibility of Streptococcus pneumoniae, Streptococcus pyogenes and Haemophilus influenzae collected from patients across the USA, in 2001-2002, as part of the PROTEKT US study." J Antimicrob Chemother 54:7-15
[Crossref] [Google Scholar] [Indexed]
- Butaye P, Cloeckaert A, Schwarz S (2003) "Mobile genes coding for efflux-mediated antimicrobial resistance in Gram-positive and Gram-negative bacteria." Int J Antimicrob Agents 22:205-210
[Crossref] [Google Scholar] [Indexed]
- Felis E, Kalka J, Sochacki A, Kowalska K, Bajkacz S, et al. (2020) "Antimicrobial pharmaceuticals in the aquatic environment - occurrence and environmental implications." Eur J Pharmacol 866: 172813
[Crossref] [Google Scholar] [Indexed]
- O'Neal L, Alvarez D, Mendizabal-Cabrera R, Ramay BM, Graham J (2020) "Community-acquired antimicrobial resistant enterobacteriaceae in central america: A one health systematic review." Int J Environ Res Public Health 17:7622
[Crossref] [Google Scholar] [Indexed]
- Bergey DH, Breed RS, Murray EGD, Hitchens AP (1939) "Bergey’s manual of determinative bacteriology, manual of determinative bacteriology”. (5th Edition), Bailliere, Tindall & Cox publisher, London.
- Ibrahim HK (2019) "Screening and sensitivity of non-lactose fermenting bacteria to antibiotics by Vitek-2 compact system." University of Thi-Qar J Sci 7:1-11
- European Committee on Antimicrobial Susceptibility, "European Committee on Antimicrobial Susceptibility Testing Breakpoint tables for interpretation of MICs and zone diameters 2015.
- Hsueh PR, Ko WC, Wu JJ, Lu JJ, Wang FD, et al. (2010) "Consensus Statement on the adherence to Clinical and Laboratory Standards Institute (CLSI) antimicrobial susceptibility testing guidelines (CLSI-2010 and CLSI-2010-update) for Enterobacteriaceae in clinical microbiology laboratories in Taiwan." J Microbiol Immunol Infect 43:452-455
[Crossref] [Google Scholar] [Indexed]
- Abat c, Rolain JM, Dubourg G, Fournier PE, Chaudet H, et al. (2017) "Evaluating the clinical burden and mortality attributable to antibiotic resistance: the disparity of empirical data and simple model estimations." Clin Infect Dis 65:58-63
[Crossref] [Google Scholar] [Indexed]
- Kahlmeter G, Giske CG, Kirn TJ, Sharp S (2019) "Point-counterpoint: differences between the European committee on antimicrobial susceptibility testing and Clinical and Laboratory standards institute recommendations for reporting antimicrobial susceptibility results." J Clin Microbiol 57:01129-1219
[Crossref] [Google Scholar] [Indexed]
- Kahlmeter G, Giske CG, Kirn TJ, Sharp S (2019) "Isolation and characterization of multiple drug resistance bacterial pathogens from waste water in hospital and non-hospital environments, Northwest Ethiopia." J Clin Microbiol 57:1-6
[Crossref] [Google Scholar] [Indexed]
- Le TH, Ng C, Chen H, Yi XZ, Koh TH, et al. (2016) "Occurrences and characterization of antibiotic-resistant bacteria and genetic determinants of hospital wastewater in a tropical country." Antimicrob Agents Chemother 60: 7449-7456
[Crossref] [Google Scholar] [Indexed]
- Dincer S, Sunduz Yigittekin E (2017) "Spreading of antibiotic resistance with wastewater." Biol Wastewater Treat Resour Recov 73:1-26
[Crossref] [Google Scholar] [Indexed]
- Fouz N, Pangesti K, Yasir M, Al-Malki A, Azhar E, et al. (2020) "The contribution of wastewater to the transmission of antimicrobial resistance in the environment: Implications of mass gathering settings." Trop Med Infect Dis 5:33
[Crossref] [Google Scholar] [Indexed]
- Harb A, Abraham S, Rusdi B, Laird T, Odea M, et al. (2019) "Molecular detection and epidemiological features of selected bacterial, viral, and parasitic enteropathogens in stool specimens from children with acute diarrhea in Thi-Qar Governorate, Iraq." Int J Environ Res Public Health 16:1573
[Crossref] [Google Scholar] [Indexed]
- Roderova M, Sedlakova M, Pudova V, Hricova K, Silova R, et al. (2016) "Occurrence of bacteria producing broad-spectrum beta-lactamases and qnr genes in hospital and urban wastewater samples." New Microbiol 39:124-33
- Rozman U, Duh D, Cimerman M, Turk S (2020) "Hospital wastewater effluent: Hot spot for antibiotic resistant bacteria." J Water Sanit Hyg Dev 10:171–178
[Crossref] [Google Scholar] [Indexed]
- Adefisoye M, Okoh A (2016) "Identification and antimicrobial resistance prevalence of pathogenic Escherichia coli strains from treated wastewater effluents in Eastern Cape, South Africa.” Microbiologyopen 5:143-51
[Crossref] [Google Scholar] [Indexed]
- Mustapha A (2019) "Detection of multidrug-resistance gram-negative bacteria from hospital sewage in North East, Nigeria." Front Environ Microbiol 5:1-7
[Crossref] [Google Scholar] [Indexed]
- Akiba M, Senba H, Otagiri H, Prabhasankar V, Taniyasu S, et al. (2015) "Impact of wastewater from different sources on the prevalence of antimicrobial-resistant Escherichia coli in sewage treatment plants in South India." Ecotoxicol Environ Saf 115:203-208
[Crossref] [Google Scholar] [Indexed]
- Asfaw T, Negash L, Kahsay A, Weldu Y (2017) "Antibiotic resistant bacteria from treated and untreated hospital wastewater at ayder referral hospital, Mekelle, North Ethiopia." Adv Microbiol 7:871-886
[Crossref] [Google Scholar] [Indexed]
- Mustafa Al Aukidy, Saeb Al Chalabi, V. P and Abstract, "Hospital Wastewater-Characteristics, Management, Treatment and Environmental Risks
- Al-Enazi MS (2016) "Evaluation of wastewater discharge from Al-Sadr teaching hospital and its impact on the Al-Khorah Channel and Shatt Al- Arab River in Basra City-Iraq." Environ Earth Sci 6:1-11
- Al-Khafaji S, Al-Rekabi W (2021) "Evaluation of the hospital’s wastewater treatment plant in basrah province." J Phys Conf Ser 1973:012033
[Crossref] [Google Scholar] [Indexed]
- Lien LTQ, Hoa NQ, Chuc NTK, Thoa NTM, Phuc HD, et al. (2016) "Antibiotics in wastewater of a rural and an urban hospital before and after wastewater treatment, and the relationship with antibiotic use-a one year study from Vietnam." Int J Environ Res Public Health 13:588
[Crossref] [Google Scholar] [Indexed]
- Rabbani M, Howlader MZH, Kabir Y (2017) "Detection of Multidrug Resistant (MDR) bacteria in untreated waste water disposals of hospitals in Dhaka City, Bangladesh." J Glob Antimicrob Resist
[Crossref] [Google Scholar] [Indexed]
- Abdulhaq A, Basode VK (2015) “Prevalence of extended-spectrum β-lactamase-producing bacteria in hospital and community sewage in Saudi Arabia." Am J Infect Control 43: 1139-1141
[Crossref] [Google Scholar] [Indexed]
- Obayiuwana A, Ogunjobi A, Yang M, Ibekwe M (2018) "Characterization of bacterial communities and their antibiotic resistance profiles in wastewaters obtained from pharmaceutical facilities in lagos and Ogun states, Nigeria." Int J Environ Res Public Health 15:1365
[Crossref] [Google Scholar] [Indexed]
- Vaz-Moreira I, Varela AR, Pereira TV, Fochat RC, Manaia CM (2016) "Multidrug resistance in quinolone-resistant gram-negative bacteria isolated from hospital effluent and the municipal wastewater treatment plant." Microb Drug Resist 22:155-163
[Crossref] [Google Scholar] [Indexed]
- Na G, Fang X, Cai Y, Ge L, Zong H, et al. (2013) "Occurrence, distribution, and bioaccumulation of antibiotics in coastal environment of Dalian, China." Mar Pollut Bull 69: 233-237
[Crossref] [Google Scholar] [Indexed]
- Bártíková H, Podlipná R, Skálová L (2016) "Veterinary drugs in the environment and their toxicity to plants." Chemosphere 144:2290-301
[Crossref] [Google Scholar] [Indexed]
- Amin DHM, Qadir C (2020) "Prevalence of Infective agents on equipment used in female cosmetic centers in Sulaymaniyah Governorate-Iraq." EurAsian J Biosci 14: 5939-5941
- Lobanovska, Mariya, Pilla, Giulia (2017) "Focus: drug development: Penicillin’s discovery and antibiotic resistance: lessons for the future?“ Yale J Biol Med 90:135-145
- Amin DMH, Guler E, Baddal B (2020) "Prevalence of Panton-Valentine leukocidin in methicillin-resistant Staphylococcus aureus clinical isolates at a university hospital in Northern Cyprus: a pilot study." BMC res notes 13: 490
[Crossref] [Google Scholar] [Indexed]
Open Access Journals
- Aquaculture & Veterinary Science
- Chemistry & Chemical Sciences
- Clinical Sciences
- Engineering
- General Science
- Genetics & Molecular Biology
- Health Care & Nursing
- Immunology & Microbiology
- Materials Science
- Mathematics & Physics
- Medical Sciences
- Neurology & Psychiatry
- Oncology & Cancer Science
- Pharmaceutical Sciences