Molecular Identification of Extended Spectrum Beta Lactamase Producing Organisms from Students in a University Community
Afunwa Ruth Asikiya1*, Nwofia Martin Chukwunonso1, Ugwu Precious Chisom1, Gbadamosi Francis Ayodele2, Ikegbune Chidozie1, Ezebialu Chinenye Uzoamaka3 and Ezemba Constance Chinyere1
1Department of Pharmaceutical Microbiology and Biotechnology, Chukwuemeka Odumegwu Ojukwu University, Igbariam, Anambra State, Nigeria
2Department of Biological Sciences, Godfrey Okoye University, Enugu State, Nigeria
3Department of Applied Microbiology and Brewing, Nnamdi Azikiwe University, Awka, Nigeria
- *Corresponding Author:
- Afunwa Ruth Asikiya
Department of Pharmaceutical Microbiology and Biotechnology,
Chukwuemeka Odumegwu Ojukwu University,
Igbariam, Anambra State,
Nigeria
Tel: 2348027341073
E-mail: drruthafunwa@yahoo.com
Received date: August 09, 2023, Manuscript No. IPJPHM-23-17678; Editor assigned date: August 11, 2023, PreQC No. IPJPHM-23-17678 (PQ); Reviewed date: August 25, 2023, QC No. IPJPHM-23-17678; Revised date: October 10, 2023, Manuscript No. IPJPHM-23-17678 (R); Published date: October 17, 2023, DOI: 10.36648/IPJPHM.9.1.001
Citation: Afunwa RA, Nwofia MC, Ugwu PC, Gbadamosi, FA, Ikegbune C, et al. (2023) Molecular Identification of Extended Spectrum Beta Lactamase Producing Organisms from Students in a University Community. J Pharm Microbiol Vol:9 No:1
Abstract
Antibiotic therapeutic failures and the surge in resistance remains a global concern. Extended Spectrum Beta Lactamase (ESBL) producing bacteria are bacteria enzymes that hydrolyze and confer resistance to newest cephalosporin antibiotics. They constitute the major mechanism of resistance especially in developing countries. Random use of antibiotics by teens and young adults for prophylaxis and treatment of infections is on the rise due largely to the “antibiotic can cure it all” mindset. This study is aimed at determining the prevalence of ESBL producing bacteria among students at Chukwuemeka Odumegwu Ojukwu university, Igbariam, Anambra state, Nigeria. The study included seventy students of both genders between ages 18-24. Clean catch midstream urine samples were collected by non-repetitive random sampling method. Seventy five isolates from the Enterobacteriaceaes were identified. The isolates were cultured on MacConkey, Cetrimide and Salmonella Shigella agar and identified using standard biochemical tests. The isolates were exposed to multi-antibiotic discs; cephalexin, levofloxacin, ofloxacin, ampicillin, augmentin, gentamicin and co-trimoxazole and phenotypically tested as ESBL producing bacteria using the Double Disc Synergy Test (DDST). Molecular identification of these isolates was also determined using forward and reverse primers of SHV, CTX and BLAOXA. The seventy five isolates obtained include; (Proteus spp. 32 (43%), E. coli 28 (37%) and Klebsiella spp. 15 (20%). Results of antimicrobial susceptibility tests for the isolates on multi antibiotic gram negative disc were interpreted using the EUCAST 2021 standard breakpoints and 75% of the isolates showed resistance to the antibiotics used. An increase in inhibition zone diameters of ≥ 5 mm confirmed ESBL production among the isolates. The results of the molecular studies confirmed the presence of BlaSHV gene after PCR and a gene size of above 1000 bp. This study identified organisms within the Enterobacteriaceaes which are listed in the WHO global priority antibiotic resistant bacteria of critical concern. The incidence of ESBL producing bacteria among the study group in the University community may be as a result of low drug knowledge and uncontrolled use of antibiotics. Antimicrobial stewardship programs are required to correct the mindset and may also break the
cycle of antibiotic abuse.
Keywords
Antibiotics; Enterobacteriaceae; Resistance; Extended Spectrum β-Lactamase (ESBLs)
Introduction
Escherichia coli, a member of the Enterobacteriaceae family, has been reported to be one of the most predominant organisms causing Urinary Tract Infections (UTIs) which are very common reasons for consultation and antibiotic prescription in current practice [1,2]. Massive and usually inappropriate use of antibiotics for treatment of UTIs generates a selective pressure that is followed by the rapid emergence and spread of multi-drug resistant bacterial strains. Nowadays, resistance of uropathogenic E. coli to many antibiotic classes is a very common finding in human medicine and is usually associated with increased medical costs, prolonged hospital stays and frequent therapeutic failure [3].
The production of Extended Spectrum β-Lactamases (ESBL) by urinary E. coli strains is a major public health concern in both hospital and community settings. These ESBL-producing strains represent a significant therapeutic challenge as they are resistant to all currently available β-lactam antibiotics but cephamycins (e.g., cefoxitin and cefotetan) and carbapenems (e.g. imipenem and ertapenem) [4].
Resistance to expanded-spectrum cephalosporins by ESBL production is mainly due to members of the TEM and SHV families of enzymes. The distribution of ESBLs has evolved to a predominance of CTX-M enzymes, mainly with E. coli as one of the major carriers of ESBL-encoding genes. Nowadays, the class-A ESBLs, TEM, SHV and CTX-M types, are the most widespread and clinically relevant [5].
Materials and Methods
Study design
The study was conducted in faculty of pharmaceutical sciences, Chukwuemeka Odumegwu Ojukwu university, Igbariam campus Anambra state from October 2021 to February 2022. Urine samples were obtained by simple random sampling method from students within the faculty. The study included 70 students of both genders between ages 18-24. The samples were collected using the clean catch midstream urine sampling technique.
Bacterial isolates
Urine samples were inoculated into 5 ml of nutrient broth and left for 24 hours. A loopful from the nutrient broth was cultured on three different selective agar; MacConkey agar, Cetrimide agar and Salmonella Shigella agar plates. The plates were incubated for 24 h at 37°C. Urine samples with positive cultures with a colony count ≥ 105 colony forming units per milliliter (CFU/mL) were selected. Out of 70 non repetitive samples included in the study, 75 isolates of Enterobacteriaceae were identified. Enterobacteriaceae isolates were identified by the standard biochemical tests including indole, oxidase test, catalase test and citrate utilization tests.
Antibiotic susceptibility testing
Disk difusion method was used for identification of antibiotic susceptibility of the Enterobacteriaceae isolates to different antibiotics according to EUCAST guidelines. The used discs were; ofloxacin (10 μg), pefloxacin (10 μg), ciprofloxacin (10 μg), amoxicillin-clavulanic acid (30 μg), gentamicin (10 μg), streptomycin (30 μg), cephalexin (10 μg), nalidixic acid (30 μg), co-trimoxazole (30 μg), ampicillin (30 μg). Resistance to three or more classes of antimicrobial agents is defined as Multiple Drug Resistance (MDR) [6].
Test for ESBL production
The isolates were tested for ESBL production using the Double Disc Synergy Test (DDST) method as previously established procedure [7]. Briefly; a combination disc (amoxicillin 20 μg and clavulanic acid 10 μg) was placed at the centre of the Petri dish and antibiotics (cefotaxime 30 μg and ceftriaxone 30 μg) were placed 15 mm apart on both sides of the plates. It was incubated at 37°C for 24 hours after which the various inhibition zone diameters were measured. Positive result is identified when the zone of inhibition is extended towards AMC (20 μg/10 μg) disc 5 mm [8,9].
DNA extraction
Genomic DNA from the isolates was extracted using Zymo research quick-DNA mini prep plus Kit (D4068). 20 mg of the bacteria cells were re-suspended in 200 ul of molecular grade water in a micro centrifuge tube. 200 μ of bio fluid and 20 ul of proteinase K were added to the re suspended cells to enable lysing of the cells. The samples were vortexed for 10-15 seconds after which, they were incubated at 55°C for 10 minutes. 420 ul of the genomic binding buffer was added to the digested samples in the tubes (containing the samples) after incubation. They were thoroughly mixed by vortexing for 15 seconds. The mixture was transferred to a Zymo-spinTM IIC-XLR column in a collection tube. After this, the tubes were placed in a micro centrifuge and centrifuged at 12,000 rpm for 1 minute. The flow through and collection tube from this process was discarded. 400 μl of DNA pre-wash buffer was added to the spin column in a new collection tube and centrifuged at 12,000 rpm for 1 min, then the flow through was discarded. 700 μl of g-DNA wash buffer was added to the spin column and centrifuged at 12,000 rpm for 1 minute, then, the flow through was discarded. Again, 200 μl of g-DNA wash buffer was added to the spin column and centrifuged at 12,000 rpm for 1 minute, then, the flow through and collection tubes were discarded. The spin column was transferred to a clean micro centrifuge tube. 50 μl of DNA Elusion buffer was added directly to the spin column matrix and incubated at room temperature for 5 minutes, then centrifuged at maximum speed for 1 minute. The resulting flow through is the DNA which was stored at 200°C.
Polymerase chain reaction for the evaluation of ITS gene region in the bacteria samples: The primers were synthesized in a DNA synthesizer (applied bio systems, UK) at InqabaBiotec company, Pretoria (South Africa). PCR was carried out in a total volume of 25 μl containing 2.0 μl of genomic DNA, 12.5 μl of 1 × PCR master mix (New England Biolabs, New York, NY, USA), 1.0 μl each of forward and reverse primer (10 mM) and 8.5 μl of H2O. The PCR protocols for BlaSHV and BlaCTX are as follows; 1 min denaturation (95°C followed by 30 cycles of 96°C for 30 s, 62°C for 30 s, and 72°C for 30 seconds and final extension of 72°C for 10 min. Conditions were identical for both assays except the annealing temperatures of BlaCTX, which is 55°C [10]. For BlaOXA, initial denaturation step at 96°C for 5 min, followed by 35 cycles consisting of denaturation at 96°C for 1 min, annealing temperature at 56°C, at 1 min, primer extension at 72°C for 1 min and final extension for 10 min. Electrophoresis of the PCR products was carried out for 30 min at 100 V on 1% (DNA samples) and 2% (for PCR amplicons) agarose gel.
| Primer | Sequence | Gene(s) | Base pair | Annealing Temperature | Reference |
|---|---|---|---|---|---|
| BlaSHV- | BlaSHV-F-GCGAAAGCCAGCTGTCGGGC (forward primer) BlaSHV-R-GATTGGCGGCGCTGTTATCGC (reverse) | Antibiotic resistance gene | >1000 BP | 62°C | Ayodele, et al. |
| BlaCTX | BlaCTX-F-GTGCAGTACCAGTAAAGTTATGG (forward) BlaCTX-R CGCAATATCATTGGTGGTGCC (reverse) | Antibiotic resistance gene | >1000 BP | 55°C | Ayodele, et al. [19] |
| BlaOXA | BlaOXA’-F-GCGCGATCTGGTTCACTCG (forward) BlaOXA-R-AGTCGACAGTTGCGCCGGC (reverse) | Antibiotic resistance gene | >1000 BP | 56°C | Iroha, et al. [20] |
Results
Out of the seventy (70) samples collected, 32 (43%) isolates of Proteus spp., 28 (37%) isolates of E. coli and 15 (20%) Kebsiella spp. were isolated.
Klebsiella spp. showed percentage resistance of >60% from ofloxacin, ciprofloxacin, augmentin, perfloxacin and ceporex while ampicillin and co-trimoxazole has 7% and 53% respectively.
Proteus spp. showed percentage resistance of >60% to ofloxacin, pefloxacin, ciprofloxacin, augmentin, gentamicin, ceporex and ampicillin while co-trimoxazole recorded 38% resistance. E. coli showed >60% resistance to ofloxacin, pefloxacin, augmentin, gentamicin, ceporex and ampicillin while ciprofloxacin and co-trimoxazole showed 53% and 31%resistance (Tables 1-3).
| Isolate | Frequency of isolate |
|---|---|
| Proteus spp. | 43% (32) |
| E.coli | 37% (28) |
| Klebsiella spp. | 20% (15) |
| Total | 100% (75) |
Table 1: Percentage frequency of isolates.
| Probable organism | OFX | PEF | CPX | AU | CN | CEP | SXT | PN |
|---|---|---|---|---|---|---|---|---|
| Proteus spp. | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| E.coli | 23 | 0 | 17 | 13 | 18 | 0 | 21 | 17 |
| Proteus spp. | 20 | 26 | 25 | 26 | 18 | 0 | 21 | 16 |
| Proteus spp. | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| E. coli | 15 | 14 | 17 | 15 | 16 | 14 | 20 | 11 |
| E. coli | 23 | 27 | 22 | 19 | 15 | 0 | 20 | 16 |
| E. coli | 20 | 16 | 24 | 15 | 16 | 0 | 17 | 16 |
| Proteus spp. | 24 | 21 | 28 | 25 | 27 | 20 | 24 | 16 |
| Proteus spp. | 12 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Proteus spp. | 33 | 31 | 24 | 16 | 28 | 16 | 26 | 0 |
| Klebsiella spp. | 26 | 20 | 22 | 22 | 26 | 20 | 22 | 20 |
| Proteus spp. | 26 | 24 | 22 | 20 | 23 | 20 | 24 | 20 |
| Proteus spp. | 30 | 30 | 28 | 16 | 20 | 0 | 16 | 0 |
| E. coli | 20 | 20 | 28 | 14 | 25 | 0 | 24 | 0 |
| Proteus spp. | 16 | 0 | 15 | 16 | 15 | 18 | 18 | 16 |
| Klebsiella spp. | 20 | 0 | 26 | 19 | 20 | 22 | 18 | 0 |
| Proteus spp. | 0 | 15 | 22 | 10 | 0 | 11 | 21 | 19 |
| Proteus spp. | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| E. coli | 26 | 22 | 26 | 10 | 30 | 20 | 28 | 0 |
| E. coli | 12 | 28 | 30 | 24 | 12 | 0 | 24 | 0 |
| E. coli | 16 | 20 | 22 | 26 | 20 | 16 | 0 | 0 |
| E. coli | 16 | 19 | 18 | 19 | 16 | 0 | 13 | 16 |
| Proteus spp. | 22 | 18 | 27 | 13 | 0 | 10 | 24 | 10 |
| E. coli | 16 | 22 | 20 | 0 | 0 | 0 | 18 | 0 |
| E. coli | 15 | 0 | 0 | 14 | 16 | 0 | 0 | 0 |
| E. coli | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Proteus spp. | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| E. coli | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| E.coli | 15 | 18 | 0 | 15 | 0 | 0 | 22 | 0 |
| Proteus spp. | 20 | 17 | 25 | 19 | 25 | 0 | 28 | 0 |
| Proteus spp. | 20 | 19 | 22 | 22 | 25 | 0 | 19 | 0 |
| E. coli | 20 | 20 | 25 | 16 | 16 | 0 | 15 | 0 |
| E. coli | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Proteus spp. | 25 | 16 | 16 | 16 | 18 | 0 | 25 | 0 |
| E. coli | 22 | 21 | 26 | 15 | 30 | 0 | 32 | 0 |
| Klebsiella spp. | 23 | 22 | 28 | 13 | 22 | 0 | 18 | 0 |
| E. coli | 19 | 21 | 28 | 19 | 16 | 0 | 18 | 0 |
| E. coli | 19 | 21 | 21 | 20 | 14 | 0 | 18 | 0 |
| E. coli | 20 | 16 | 29 | 20 | 21 | 0 | 25 | 0 |
| Proteus spp. | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| E. coli | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Proteus spp. | 18 | 0 | 20 | 18 | 16 | 0 | 0 | 0 |
| E. coli | 19 | 18 | 27 | 19 | 23 | 0 | 16 | 0 |
| E. coli | 0 | 20 | 0 | 0 | 27 | 0 | 0 | 0 |
| E. coli | 19 | 25 | 20 | 0 | 14 | 0 | 12 | 14 |
| Proteus spp. | 19 | 16 | 18 | 12 | 10 | 0 | 20 | 12 |
| E.coli | 20 | 19 | 20 | 13 | 23 | 0 | 27 | 16 |
| Klebsiella spp. | 20 | 26 | 0 | 14 | 0 | 12 | 20 | 0 |
| E. coli | 0 | 15 | 16 | 14 | 13 | 0 | 21 | 16 |
| Klebsiella spp. | 19 | 25 | 18 | 22 | 20 | 0 | 18 | 0 |
| Proteus spp. | 30 | 30 | 18 | 20 | 0 | 28 | 30 | 0 |
| Klebsiella spp. | 31 | 18 | 20 | 18 | 0 | 12 | 32 | 16 |
| Klebsiella spp. | 26 | 20 | 26 | 15 | 30 | 20 | 24 | 0 |
| Klebsiella spp. | 28 | 25 | 26 | 20 | 26 | 0 | 20 | 23 |
| Proteus spp. | 17 | 15 | 13 | 0 | 0 | 0 | 16 | 17 |
| E. coli | 17 | 0 | 25 | 0 | 0 | 0 | 15 | 0 |
| Proteus spp. | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Proteus spp. | 21 | 20 | 21 | 20 | 18 | 0 | 20 | 21 |
| Proteus spp. | 17 | 13 | 0 | 0 | 21 | 0 | 23 | 0 |
| Proteus spp. | 26 | 18 | 19 | 23 | 14 | 0 | 27 | 18 |
| E. coli | 0 | 0 | 25 | 14 | 16 | 0 | 32 | 15 |
| Proteus spp. | 0 | 16 | 0 | 14 | 0 | 0 | 0 | 14 |
| Klebsiella spp. | 14 | 18 | 10 | 0 | 0 | 0 | 18 | 0 |
| Klebsiella spp. | 17 | 16 | 19 | 14 | 20 | 0 | 21 | 15 |
| Klebsiella spp. | 15 | 18 | 20 | 0 | 15 | 0 | 22 | 16 |
| Proteus spp. | 0 | 0 | 10 | 13 | 0 | 0 | 21 | 13 |
| Proteus spp. | 0 | 0 | 15 | 0 | 0 | 0 | 0 | 0 |
| Proteus spp. | 0 | 0 | 0 | 0 | 16 | 0 | 0 | 0 |
| Klebsiella spp. | 0 | 17 | 0 | 0 | 0 | 0 | 0 | 0 |
| Klebsiella spp. | 22 | 25 | 20 | 21 | 20 | 0 | 25 | 21 |
| Klebsiella spp. | 0 | 16 | 15 | 0 | 8 | 0 | 17 | 21 |
| Klebsiella spp. | 0 | 0 | 13 | 0 | 0 | 0 | 18 | 0 |
| Proteus spp. | 0 | 16 | 18 | 20 | 0 | 0 | 21 | 15 |
| Proteus spp. | 12 | 14 | 0 | 0 | 12 | 0 | 12 | 0 |
| Proteus spp. | 12 | 10 | 15 | 12 | 13 | 0 | 22 | 15 |
| Note: OFX–Ofloxacin; PEF–Levofloxacin; CPX–Ciprofloxacin; AU-Augmentin; CN–Gentamicin; CEP–Ceporex; SXT–Co-trimoxazole; PN–Ampicilin. | ||||||||
Table 2: Antibiotics susceptibility test result (inhibition zone diameter measured in mm).
| Isolate | IZD of CR | IZD of CF+AMC | IZD of CF | IZD of CR+AMC | Remark |
|---|---|---|---|---|---|
| Klebsiella spp. | 7 | 20 | 9 | 23 | + |
| Klebsiella spp. | 10 | 25 | 10 | 26 | + |
| E. coli | 9 | 25 | 11 | 27 | + |
| Proteus spp. | 3 | 0 | 4 | 0 | - |
| Proteus spp. | 0 | 0 | 0 | 0 | - |
| Proteus spp. | 12 | 26 | 11 | 21 | + |
| E. coli | 13 | 30 | 11 | 24 | + |
| Proteus spp. | 7 | 23 | 5 | 22 | + |
| E.coli | 4 | 0 | 1 | 0 | - |
| E.coli | 9 | 22 | 8 | 21 | + |
| Proteus spp. | 0 | 0 | 0 | 0 | - |
| Proteus spp. | 3 | 0 | 4 | 0 | - |
| E. coli | 12 | 0 | 11 | 0 | - |
| E. coli | 8 | 20 | 10 | 22 | + |
| Proteus spp. | 14 | 24 | 12 | 25 | + |
| Klebsiella spp. | 11 | 25 | 9 | 23 | + |
| Proteus spp. | 6 | 20 | 7 | 16 | + |
| Proteus spp. | 6 | 19 | 5 | 16 | + |
| Proteus spp. | 7 | 0 | 8 | 0 | - |
| Proteus spp. | 0 | 0 | 0 | 0 | - |
| E. coli | 12 | 0 | 0 | 0 | - |
| E. coli | 14 | 0 | 10 | 0 | - |
| Klebsiella spp. | 10 | 0 | 10 | 0 | - |
| Klebsiella spp. | 10 | 0 | 10 | 0 | - |
| Klebsiella spp. | 20 | 0 | 19 | 0 | - |
| Proteus spp. | 0 | 0 | 0 | 0 | - |
| Klebsiella spp. | 0 | 0 | 0 | 0 | - |
| Proteus spp. | 20 | 0 | 0 | 0 | - |
| E. coli | 20 | 0 | 17 | 0 | - |
| Proteus spp. | 19 | 0 | 14 | 0 | - |
| Note: Positive (+) This means that the resistance is mediated via the beta-lactamse enzyme. Negative (-) This means that the resistance is not mediated via beta-lactamase enzyme. IZD-Inhibition Zone Diameter; CF-Cefotaxime; CR-Ceftriaxone; AMC-Augmentin (Amoxicillin-clavulanic acid) |
|||||
Table 3: Phenotypic confirmation of ESBL production using the Double Disc Synergy Test (DDST) Inhibition Zone Diametre (IZD).
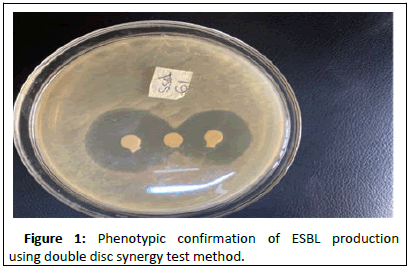
Result of the phenotypic double disc synergy test is shown in Figure 1 below. The result showed positive for twelve (12) isolates. Of the twelve isoltes identified as ESBL producers, 5 (41.67%) were Proteus spp., 4 (33.33%) were E. coli and 3 (25%) were Klebsiella spp. (Figures 1 and 2).
Discussion
The amount of infections caused by Extended Spectrum Beta Lactamase producing Enterobacteriaceae (ESBL-E) has increased exponentially around the world and this infections are associated with higher morbidity and mortality rates. Among various reasons, this problem has been linked with improper use of antibiotics during these years [11]. Enterobacteriaceae are the most common pathogens causing Urinary Tract Infections (UTIs). Increasing rates of antimicrobial resistance among Enterobacteriaceae strains decreases the option of empiric treatment of these infections [12].
Results of antimicrobial susceptibility test for the isolates respectively using the multi-antibiotic disc (gram-negative) were interpreted using the EUCAST 2021 standard breakpoints. For E. coli, our study indicated higher percentage resistance of isolates to floro quinolones. This is in contradiction with what was reported by Ugwu, et al., which recorded susceptibility of E. coli to fluoro quinolones (75.86%). This could be explained that E. coli around Igbariam axis is expressing higher resistance gene towards ciprofloxacin.
For Proteus spp., 84% of the isolates were resistant to cephalexin. Levofloxacin and ofloxacin had the same resistance pattern of 78% resistance. The isolates when tested against ciprofloxacin had 71% resistance, 16% intermediate and 13% susceptibility while ampicillin and Augmentin had 69% and 71% resistance respectively. Gentamicin had 69% resistance and 31% susceptibility while co-trimoxazole had 62% susceptibility and 38% resistance. Similar data regarding resistance against ampicillin and amoxicillin clavulanate were reported by Gonzalez, et al. and Pathak, et al. [4,13].
For Klebsiella spp., ofloxacin, ciprofloxacin and Augmentin showed 73% resistance while levofloxacin and cephalexin showed 67% resistance to the isolate. The isolate when tested against gentamicin 47% resistance and 53% susceptibility while ampicillin had 53% resistance and 47% susceptibility. Co-trimoxazole showed improved action against the isolate with 93% susceptibility and 7% resistance. Our data is consistent with those obtained by Pathek, et al., and Teklu, et al., showing high resistance of Klebsiella spp. to floro quinolones and augmentin [14].
Using the double disc synergy test as described by Rawat, et al., our findings revealed that out of 30 isolates tested, 40% were ESBL positive having a zone of inhibition of ≥ 5 mm. This is in agreement with studies performed by Tanko, et al., 50.8% and Abdelghani, et al., 46% [15]. Also, Afunwa, et al., reported that the community prevalence of ESBL in Southeastern Nigeria were placed at 4.4% which shows a wide margin from our findings. This can be attributed to the growing trend of inappropriate consumption of wide spectrum antibiotics over the years. Another interesting fact is that young people are habitual consumers of hamburgers/shawarma/pizza (pork, beef, and chicken), foods in which a high prevalence of these ESBL bacteria has been found [16]. Mandal, et al., and Nwankwo, et al., reported prevalence of 28.46% and 12.8% respectively. The variations may be due to the different sources of samples used in the studies [17].
The prevalence of ESBL in developing countries like Nigeria is higher than in developed countries because of the absence of antimicrobial stewardship program, indiscriminate use of antibiotics and lack of adequate antimicrobial resistance surveillance [18-20].
Conclusion
The findings from this study show a 40% prevalence of ESBL producing organism among tested students within the University. This can be as a result of uncontrolled use of antibiotics and poor level of enlightenment about antimicrobial stewardship programs. This study provides data to monitor the surge of ESBL producing isolates amongst the general population in order to reduce the economic impact, improve health and avoid deaths resulting from antibiotics resistance.
References
- Adesoji AT, Ogunjobi AA (2016) Detection of extended spectrum beta-lactamases resistance genes among bacteria isolated from selected drinking water distribution channels in Southwestern Nigeria. Biomed Res Int 2016:1-9
- Afunwa RA, Esimone CO, Iroha RI, Odimegwu DC (2011) Antimicrobial resistance status and prevalence rates of extended spectrum beta lactamase producers isolated from a mixed human population. Bosn J Basic Med Sci 11:91-96
[Crossref]
- de Angelis G, del Giacomo P, Posteraro B, Sanguinetti M, Tumbarello M (2010) Molecular mechanisms, epidemiology, and clinical importance of β-lactam resistance in Enterobacteriaceae. Int J Mol Sci 21:5090
[Crossref] [Google Scholar] [PubMed]
- Gonzalez D, Gallagher E, Zuniga T, Leiva J, Vitas AI (2019) Prevalence and characterization of β lactamase producing Enterobacteriaceae in healthy human carriers. Int Microbiol 23:171-177
- Iroha IR, Mohammed ID, Moses IB, Ngwu NJ, Uzoeto HO, et al. (2022) Molecular characterization of enterobacteriacea isolated from gingivitis and periodontitis patients and the antimicrobial activity of mouth wash agents. Scientific African 15:e01106
- Jacoby GA, Medeiros AA (1991) More extended-spectrum beta lactamases. Antimicrob Agents Chemother 35:1697-1704
[Crossref] [Google Scholar] [PubMed]
- Lagadinou M, Onisor MO, Rigas A, Musetescu DV, Gkentzi D, et al. (2020) Antimicrobial properties on non-antibiotic drugs in the era of increased bacterial resistance. Antibiotics 9:107
[Crossref] [Google Scholar] [PubMed]
- Magiorakos AP, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, et al. (2012) Multidrug resistant, extensively drug resistant and drug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect 18:268-281
[Crossref] [Google Scholar] [PubMed]
- Mandal DK, Sah SK, Mishra SK, Sharma S, Kattel HP, et al. (2020) Carriage of extended-spectrum-β-lactamase-and AmpC-β-lactamase-producing Enterobacteriaceae (ESBL-PE) in Healthy Community and Outpatient Department (OPD) patients in Nepal. Can J Infect Dis Med Microbiol 2020:5154217
- Mohamed ES, Khairy RM, Abdelrahim SS (2020) Prevalence and molecular characteristics of ESBL and AmpC β-lactamase producing Enterobacteriaceae strains isolated from UTIs in Egypt. Antimicrob Resist Infect Control 9:1-9
[Crossref] [Google Scholar] [PubMed]
- Musa BM, Imam H, Lendel A, Abdulkadir I, Gumi HS, et al. The burden of extended-spectrum β-lactamase-producing Enterobacteriaceae in Nigeria: A systematic review and meta-analysis. Trans R Soc Trop Med Hyg 114:241-248
[Crossref] [Google Scholar] [PubMed]
- Nwankwo EO, Magaji NS, Tijjani J (2015) Antibiotic susceptibility pattern of Extended Spectrum Betalactamase (ESBL) producers and other bacterial pathogens in Kano, Nigeria. Trop J Pharm 14:1273-1278
- Pathak S, Sharma M, Srivastava P (2013) Gram negative Bacilli and further molecular Escherichia coli and Klebsiella spp. J Clin Diagnostic Res 7:21732177
- Rosantia S, Higa T, Yagi N, Tokunaga T, Higa S, et al. (2020) Characterization of CTX-M-type-Extended-Spectrum Beta-Lactamase (ESBL)-producing Enterobacteriaceae isolated from Indonesian undergraduate medical students of a university in Surabaya, Indonesia. J Infect Chemother 26:575-581.
[Crossref] [Google Scholar] [PubMed]
- Tanko N, Bolaji RO, Olayinka AT, Olayinka BO (2020) A systematic review on the prevalence of extended-spectrum beta lactamase-producing gram-negative bacteria in Nigeria. J Glob Antimicrob Resist 22:488-496
[Crossref] [Google Scholar] [PubMed]
- Brolund A, Sandegren L (2016) Characterization of ESBL disseminating plasmids. Infect Dis 48:18-25
[Crossref] [Google Scholar] [PubMed]
- Teklu DS, Negeri AA, Legese MH, Bedada TL, Woldemariam HK, et al. (2019) Extended-spectrum beta-lactamase production and multi-drug resistance among Enterobacteriaceae isolated in Addis Ababa, Ethiopia. Antimicrob Resist Infect Control 8:1-2
[Crossref] [Google Scholar] [PubMed]
- Ugwu MC, Shariff M, Nnajide CM, Beri K, Okezie UM, et al. (2020) Phenotypic and molecular characterization of β-lactamases among Enterobacterial uropathogens in Southeastern Nigeria. Can J Infect Dis Med Microbiol 2020:5843904
[Crossref]
- Adesoji AT, Ogunjobi AA (2016) Detection of extended spectrum beta-lactamases resistance genes among bacteria isolated from selected drinking water distribution channels in Southwestern Nigeria. BioMed Res Int 2016:9
[Crossrf] [Google Scholar] [PubMed]
- Iroha IR, Mohammed ID, Moses IB, Ngwu NJ, Uzoeto HO, et al. (2022) Molecular characterization of Enterobacteriacea isolated from gingivitis and periodontitis patients and the antimicrobial activity of mouth wash agents. Sci Afr 15:e01106
Open Access Journals
- Aquaculture & Veterinary Science
- Chemistry & Chemical Sciences
- Clinical Sciences
- Engineering
- General Science
- Genetics & Molecular Biology
- Health Care & Nursing
- Immunology & Microbiology
- Materials Science
- Mathematics & Physics
- Medical Sciences
- Neurology & Psychiatry
- Oncology & Cancer Science
- Pharmaceutical Sciences